

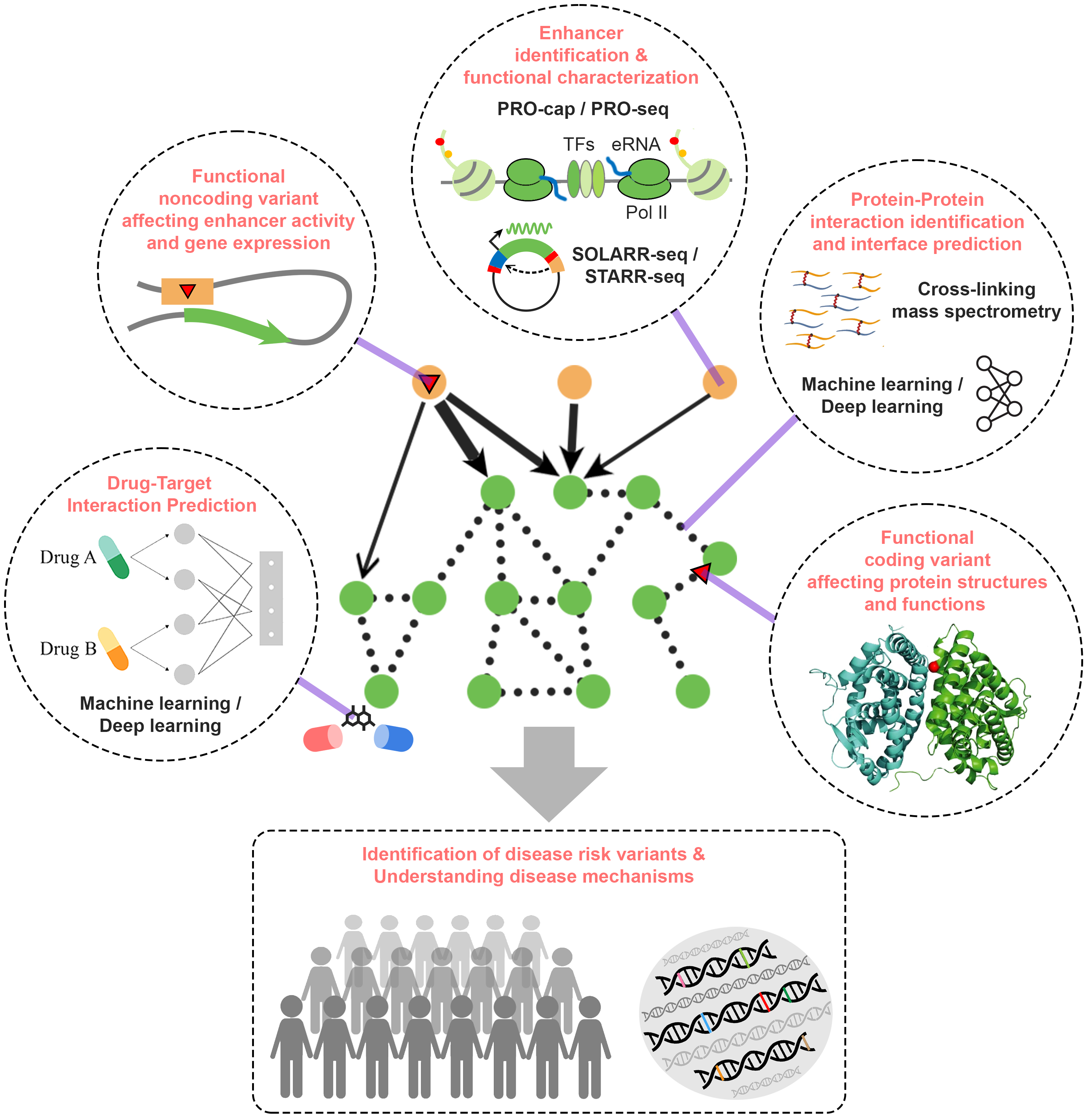
The Yu group performs research in the broad areas of Network Systems Biology. We use integrated computational-experimental systems biology approaches to determine protein interactions and complex structures on the scale of the whole cell. In particular, we focus on protein-protein and gene regulatory networks and seek to understand how such intricate systems evolve and how their perturbations lead to human diseases, especially autism spectrum disorder and cancer. Towards these goals, Dr. Haiyuan Yu led the effort to develop comprehensivethe concept of “3D structurally-resolved interactome networks”, where we integrate multi-scale structural modeling, machine learning, and high-throughput genomics/proteomics experiments to determine protein interactions and their binding interfaces on the whole proteome scale. We have developed innovative cross-linking mass spectrometry (XL-MS) and quantitative proteomics approaches to dissect native protein complexes with structural details at whole proteome scale. Furthermore, in close collaboration with Dr. John Lis and his group, we demonstrated that enhancer RNA (eRNA) transcription detected by the novel PRO-cap assay is a critical mark to define active enhancers with high resolution genome-wide. Mutations at key sites within these enhancers can affect eRNA transcription and thus significantly impact their enhancer activities. The Yu group has established many cutting-edge experimental methodologies in structural proteomics, mass spectrometry, and functional genomics, and built innovative analysis pipelines using machine learning (deep learning), graph theory, and rigorous statistical models in these areas.
Dapeng X et al. 2024
Structurally-informed human interactome reveals proteome-wide perturbations by disease mutations
Nature Biotechnology
Zhou, Liu, & Gupta et al. 2023
A comprehensive SARS-CoV-2-human protein-protein interactome reveals COVID-19 pathobiology and potential host therapeutic targets
Nature Biotechnology
Yao L et al. 2022
A comparison of experimental assays and analytical methods for genome-wide identification of active enhancers
Nature Biotechnology
Meyer MJ et al. 2018
Interactome INSIDER: a structural interactome browser for genomic studies
Nature Methods
Currently, we have open positions at all levels (e.g., post-docs, graduate students, undergrads, technicians). Details
